%matplotlib inlineplot_comparison_under_sampling
ref: imbalaced-learn
Compare under-sampling samplers
The following example attends to make a qualitative comparison between the different under-sampling algorithms available in the imbalanced-learn package.
# Authors: Guillaume Lemaitre <g.lemaitre58@gmail.com>
# License: MITprint(__doc__)
import seaborn as sns
sns.set_context("poster")Automatically created module for IPython interactive environmentThe following function will be used to create toy dataset. It uses the :func:~sklearn.datasets.make_classification from scikit-learn but fixing some parameters.
from sklearn.datasets import make_classification
def create_dataset(
n_samples=1000,
weights=(0.01, 0.01, 0.98),
n_classes=3,
class_sep=0.8,
n_clusters=1,
):
return make_classification(
n_samples=n_samples,
n_features=2,
n_informative=2,
n_redundant=0,
n_repeated=0,
n_classes=n_classes,
n_clusters_per_class=n_clusters,
weights=list(weights),
class_sep=class_sep,
random_state=0,
)The following function will be used to plot the sample space after resampling to illustrate the specificities of an algorithm.
def plot_resampling(X, y, sampler, ax, title=None):
X_res, y_res = sampler.fit_resample(X, y)
ax.scatter(X_res[:, 0], X_res[:, 1], c=y_res, alpha=0.8, edgecolor="k")
if title is None:
title = f"Resampling with {sampler.__class__.__name__}"
ax.set_title(title)
sns.despine(ax=ax, offset=10)The following function will be used to plot the decision function of a classifier given some data.
import numpy as np
def plot_decision_function(X, y, clf, ax, title=None):
plot_step = 0.02
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(
np.arange(x_min, x_max, plot_step), np.arange(y_min, y_max, plot_step)
)
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
ax.contourf(xx, yy, Z, alpha=0.4)
ax.scatter(X[:, 0], X[:, 1], alpha=0.8, c=y, edgecolor="k")
if title is not None:
ax.set_title(title)from sklearn.linear_model import LogisticRegression
clf = LogisticRegression()Prototype generation: under-sampling by generating new samples
:class:~imblearn.under_sampling.ClusterCentroids under-samples by replacing the original samples by the centroids of the cluster found.
import matplotlib.pyplot as plt
from sklearn.cluster import MiniBatchKMeans
from imblearn import FunctionSampler
from imblearn.pipeline import make_pipeline
from imblearn.under_sampling import ClusterCentroids
X, y = create_dataset(n_samples=400, weights=(0.05, 0.15, 0.8), class_sep=0.8)
samplers = {
FunctionSampler(), # identity resampler
ClusterCentroids(
estimator=MiniBatchKMeans(n_init=1, random_state=0), random_state=0
),
}
fig, axs = plt.subplots(nrows=2, ncols=2, figsize=(15, 15))
for ax, sampler in zip(axs, samplers):
model = make_pipeline(sampler, clf).fit(X, y)
plot_decision_function(
X, y, model, ax[0], title=f"Decision function with {sampler.__class__.__name__}"
)
plot_resampling(X, y, sampler, ax[1])
fig.tight_layout()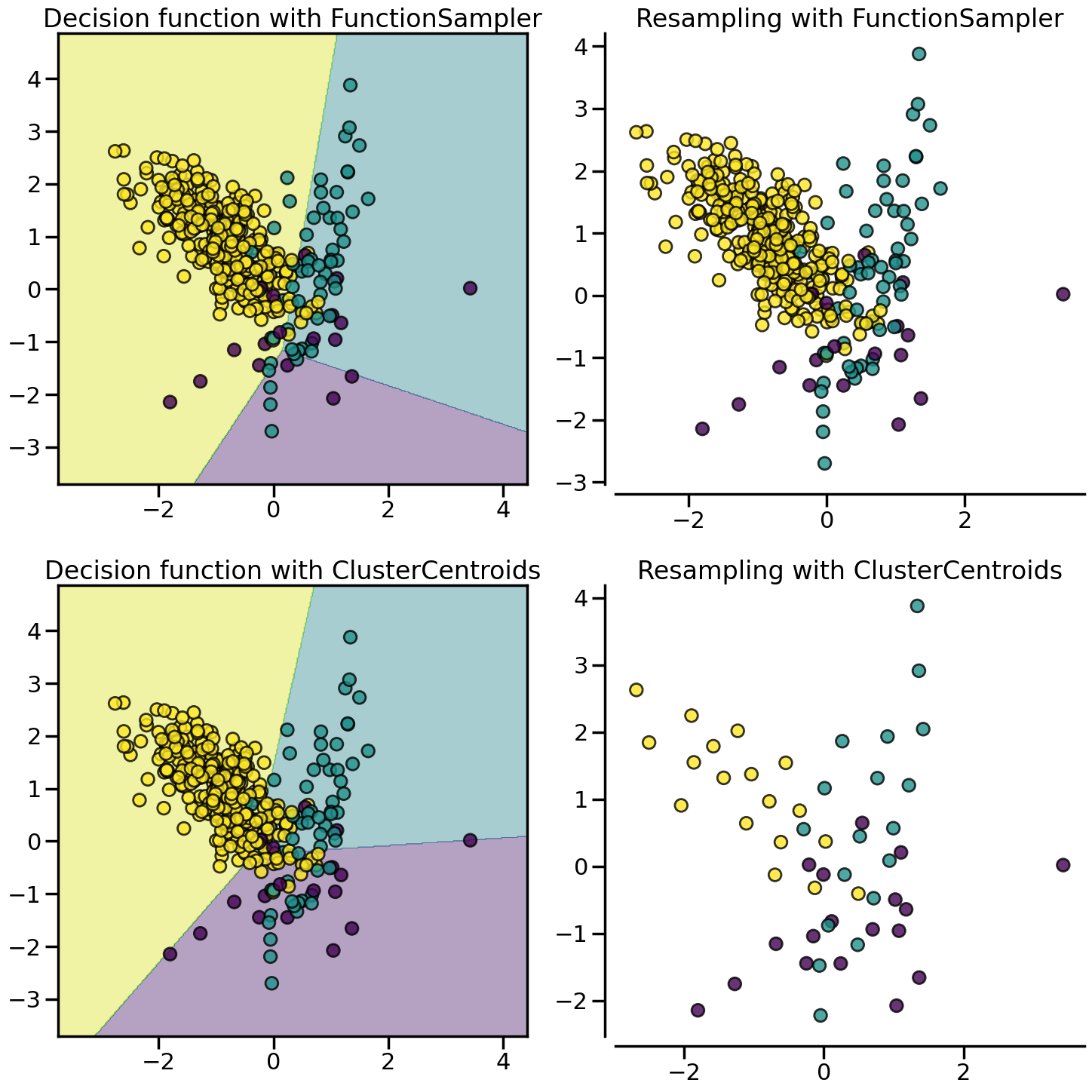
Prototype selection: under-sampling by selecting existing samples
The algorithm performing prototype selection can be subdivided into two groups: (i) the controlled under-sampling methods and (ii) the cleaning under-sampling methods.
With the controlled under-sampling methods, the number of samples to be selected can be specified. :class:~imblearn.under_sampling.RandomUnderSampler is the most naive way of performing such selection by randomly selecting a given number of samples by the targetted class.
from imblearn.under_sampling import RandomUnderSampler
X, y = create_dataset(n_samples=400, weights=(0.05, 0.15, 0.8), class_sep=0.8)
samplers = {
FunctionSampler(), # identity resampler
RandomUnderSampler(random_state=0),
}
fig, axs = plt.subplots(nrows=2, ncols=2, figsize=(15, 15))
for ax, sampler in zip(axs, samplers):
model = make_pipeline(sampler, clf).fit(X, y)
plot_decision_function(
X, y, model, ax[0], title=f"Decision function with {sampler.__class__.__name__}"
)
plot_resampling(X, y, sampler, ax[1])
fig.tight_layout()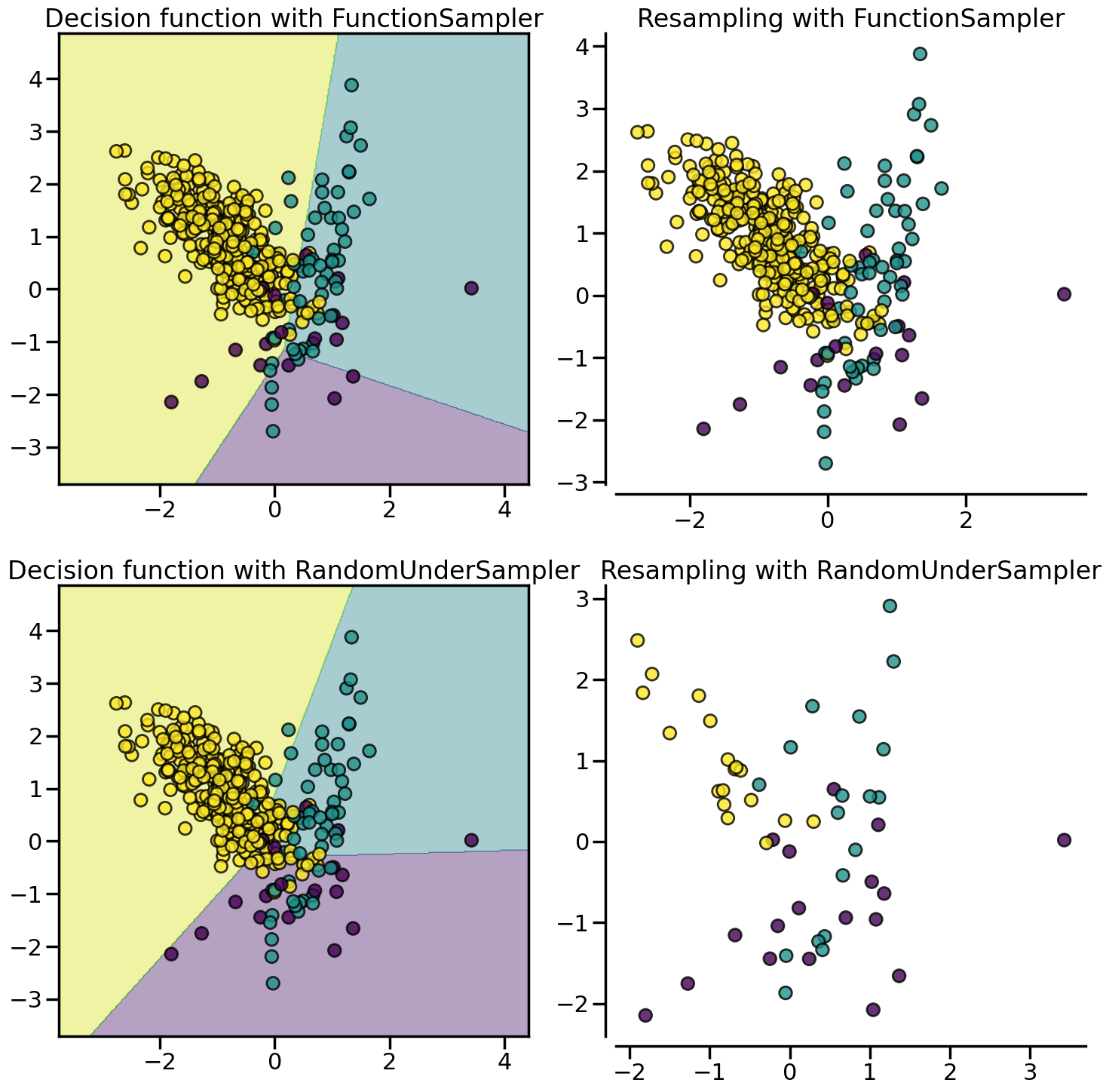
:class:~imblearn.under_sampling.NearMiss algorithms implement some heuristic rules in order to select samples. NearMiss-1 selects samples from the majority class for which the average distance of the \(k\)` nearest samples of the minority class is the smallest. NearMiss-2 selects the samples from the majority class for which the average distance to the farthest samples of the negative class is the smallest. NearMiss-3 is a 2-step algorithm: first, for each minority sample, their \(m\) nearest-neighbors will be kept; then, the majority samples selected are the on for which the average distance to the \(k\) nearest neighbors is the largest.
from imblearn.under_sampling import NearMiss
X, y = create_dataset(n_samples=1000, weights=(0.05, 0.15, 0.8), class_sep=1.5)
samplers = [NearMiss(version=1), NearMiss(version=2), NearMiss(version=3)]
fig, axs = plt.subplots(nrows=3, ncols=2, figsize=(15, 25))
for ax, sampler in zip(axs, samplers):
model = make_pipeline(sampler, clf).fit(X, y)
plot_decision_function(
X,
y,
model,
ax[0],
title=f"Decision function for {sampler.__class__.__name__}-{sampler.version}",
)
plot_resampling(
X,
y,
sampler,
ax[1],
title=f"Resampling using {sampler.__class__.__name__}-{sampler.version}",
)
fig.tight_layout()/home/coco/anaconda3/envs/py38/lib/python3.8/site-packages/imblearn/under_sampling/_prototype_selection/_nearmiss.py:203: UserWarning: The number of the samples to be selected is larger than the number of samples available. The balancing ratio cannot be ensure and all samples will be returned.
warnings.warn(
/home/coco/anaconda3/envs/py38/lib/python3.8/site-packages/imblearn/under_sampling/_prototype_selection/_nearmiss.py:203: UserWarning: The number of the samples to be selected is larger than the number of samples available. The balancing ratio cannot be ensure and all samples will be returned.
warnings.warn(
/home/coco/anaconda3/envs/py38/lib/python3.8/site-packages/imblearn/under_sampling/_prototype_selection/_nearmiss.py:203: UserWarning: The number of the samples to be selected is larger than the number of samples available. The balancing ratio cannot be ensure and all samples will be returned.
warnings.warn(
/home/coco/anaconda3/envs/py38/lib/python3.8/site-packages/imblearn/under_sampling/_prototype_selection/_nearmiss.py:203: UserWarning: The number of the samples to be selected is larger than the number of samples available. The balancing ratio cannot be ensure and all samples will be returned.
warnings.warn(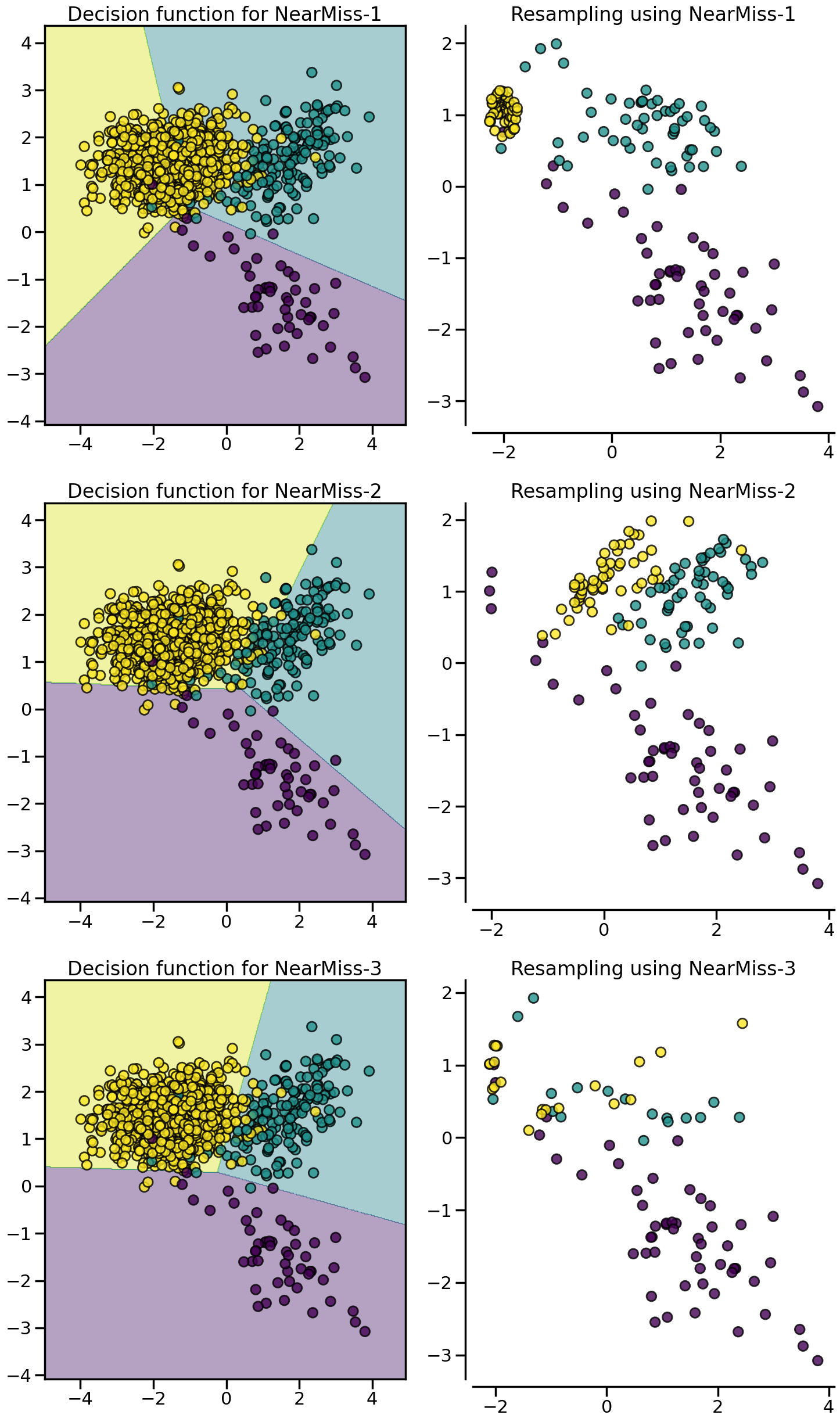
:class:~imblearn.under_sampling.EditedNearestNeighbours removes samples of the majority class for which their class differ from the one of their nearest-neighbors. This sieve can be repeated which is the principle of the :class:~imblearn.under_sampling.RepeatedEditedNearestNeighbours. :class:~imblearn.under_sampling.AllKNN is slightly different from the :class:~imblearn.under_sampling.RepeatedEditedNearestNeighbours by changing the \(k\) parameter of the internal nearest neighors algorithm, increasing it at each iteration.
from imblearn.under_sampling import (
AllKNN,
EditedNearestNeighbours,
RepeatedEditedNearestNeighbours,
)
X, y = create_dataset(n_samples=500, weights=(0.2, 0.3, 0.5), class_sep=0.8)
samplers = [
EditedNearestNeighbours(),
RepeatedEditedNearestNeighbours(),
AllKNN(allow_minority=True),
]
fig, axs = plt.subplots(3, 2, figsize=(15, 25))
for ax, sampler in zip(axs, samplers):
model = make_pipeline(sampler, clf).fit(X, y)
plot_decision_function(
X, y, clf, ax[0], title=f"Decision function for \n{sampler.__class__.__name__}"
)
plot_resampling(
X, y, sampler, ax[1], title=f"Resampling using \n{sampler.__class__.__name__}"
)
fig.tight_layout()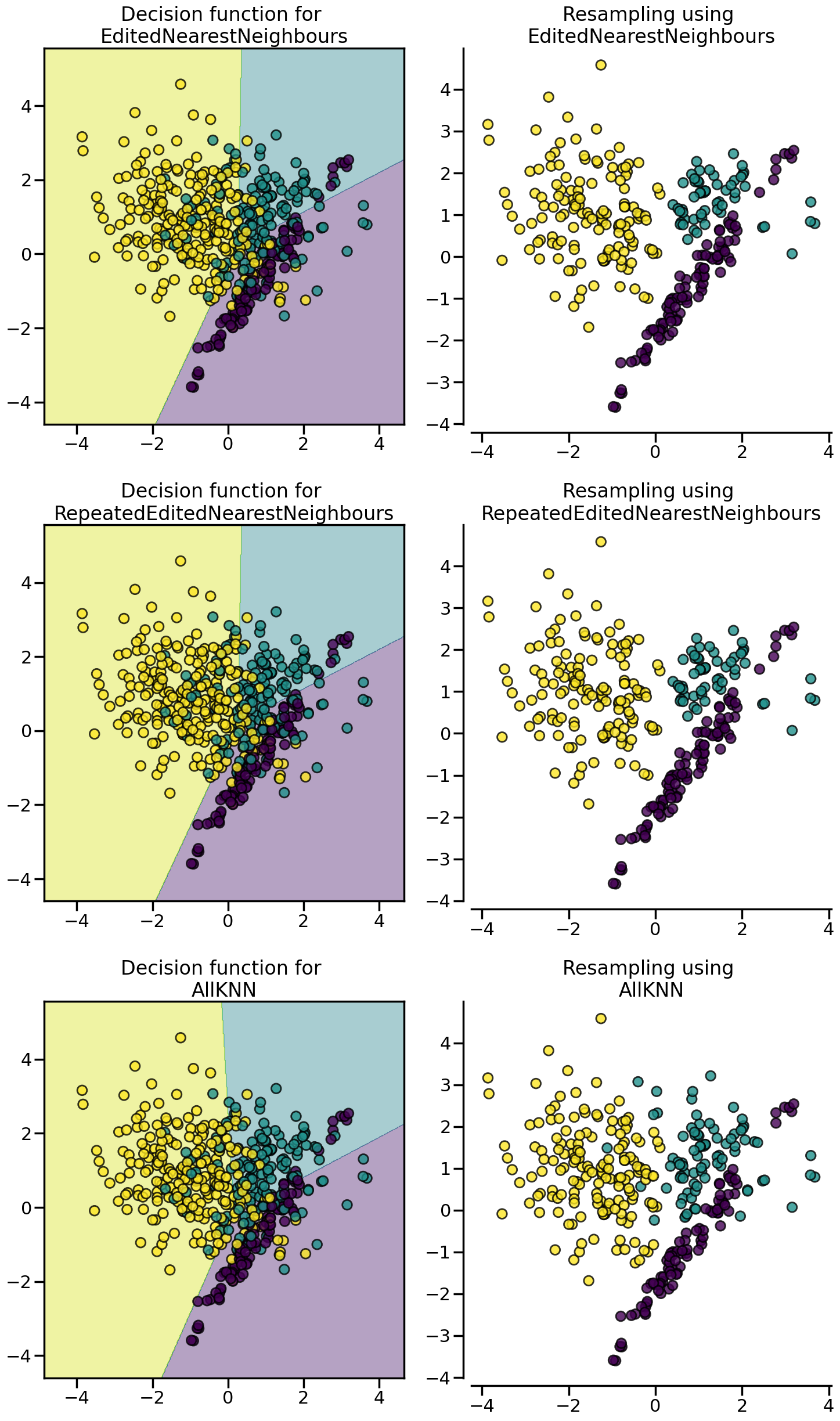
:class:~imblearn.under_sampling.CondensedNearestNeighbour makes use of a 1-NN to iteratively decide if a sample should be kept in a dataset or not. The issue is that :class:~imblearn.under_sampling.CondensedNearestNeighbour is sensitive to noise by preserving the noisy samples. :class:~imblearn.under_sampling.OneSidedSelection also used the 1-NN and use :class:~imblearn.under_sampling.TomekLinks to remove the samples considered noisy. The :class:~imblearn.under_sampling.NeighbourhoodCleaningRule use a :class:~imblearn.under_sampling.EditedNearestNeighbours to remove some sample. Additionally, they use a 3 nearest-neighbors to remove samples which do not agree with this rule.
from imblearn.under_sampling import (
CondensedNearestNeighbour,
NeighbourhoodCleaningRule,
OneSidedSelection,
)
X, y = create_dataset(n_samples=500, weights=(0.2, 0.3, 0.5), class_sep=0.8)
fig, axs = plt.subplots(nrows=3, ncols=2, figsize=(15, 25))
samplers = [
CondensedNearestNeighbour(random_state=0),
OneSidedSelection(random_state=0),
NeighbourhoodCleaningRule(),
]
for ax, sampler in zip(axs, samplers):
model = make_pipeline(sampler, clf).fit(X, y)
plot_decision_function(
X, y, clf, ax[0], title=f"Decision function for \n{sampler.__class__.__name__}"
)
plot_resampling(
X, y, sampler, ax[1], title=f"Resampling using \n{sampler.__class__.__name__}"
)
fig.tight_layout()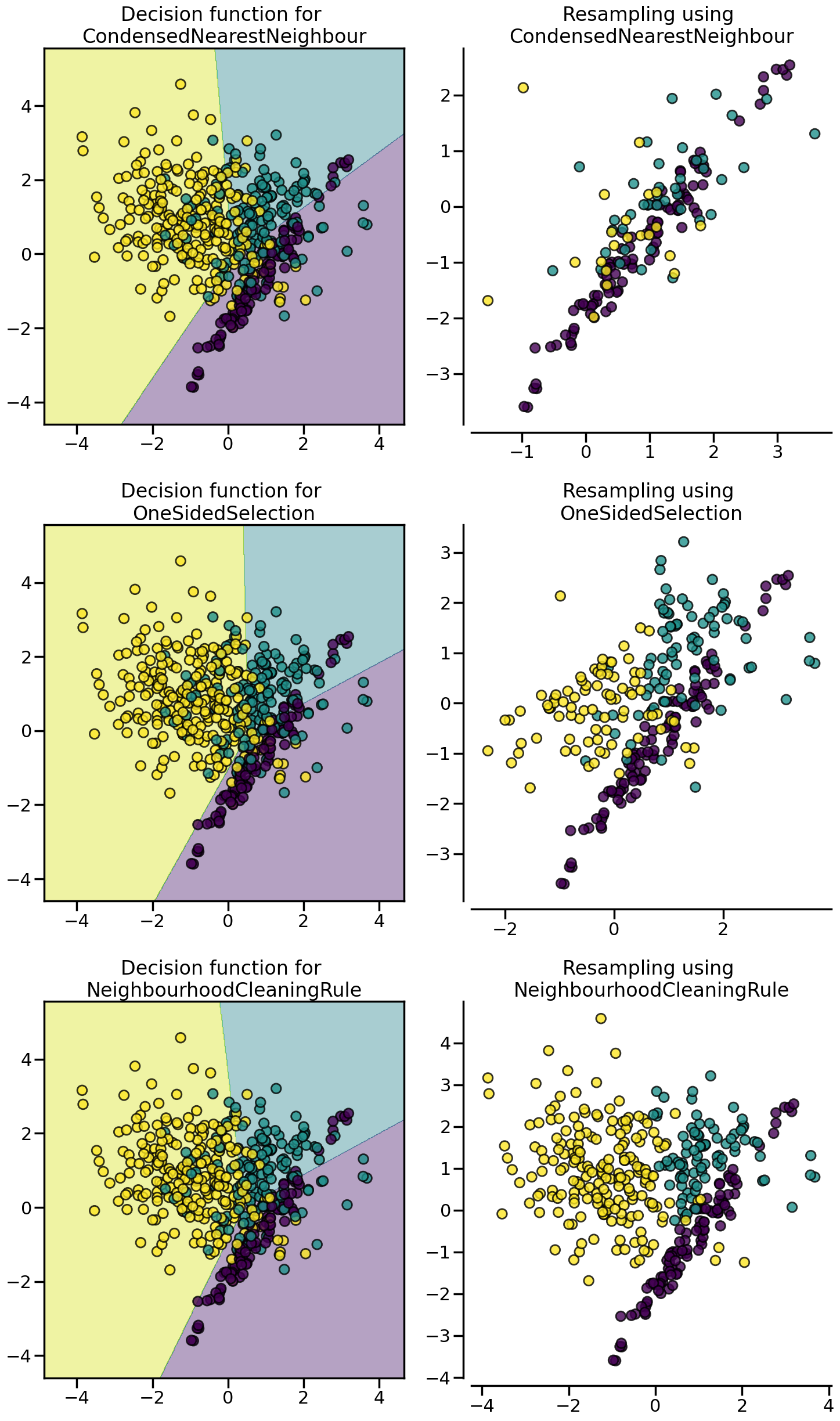
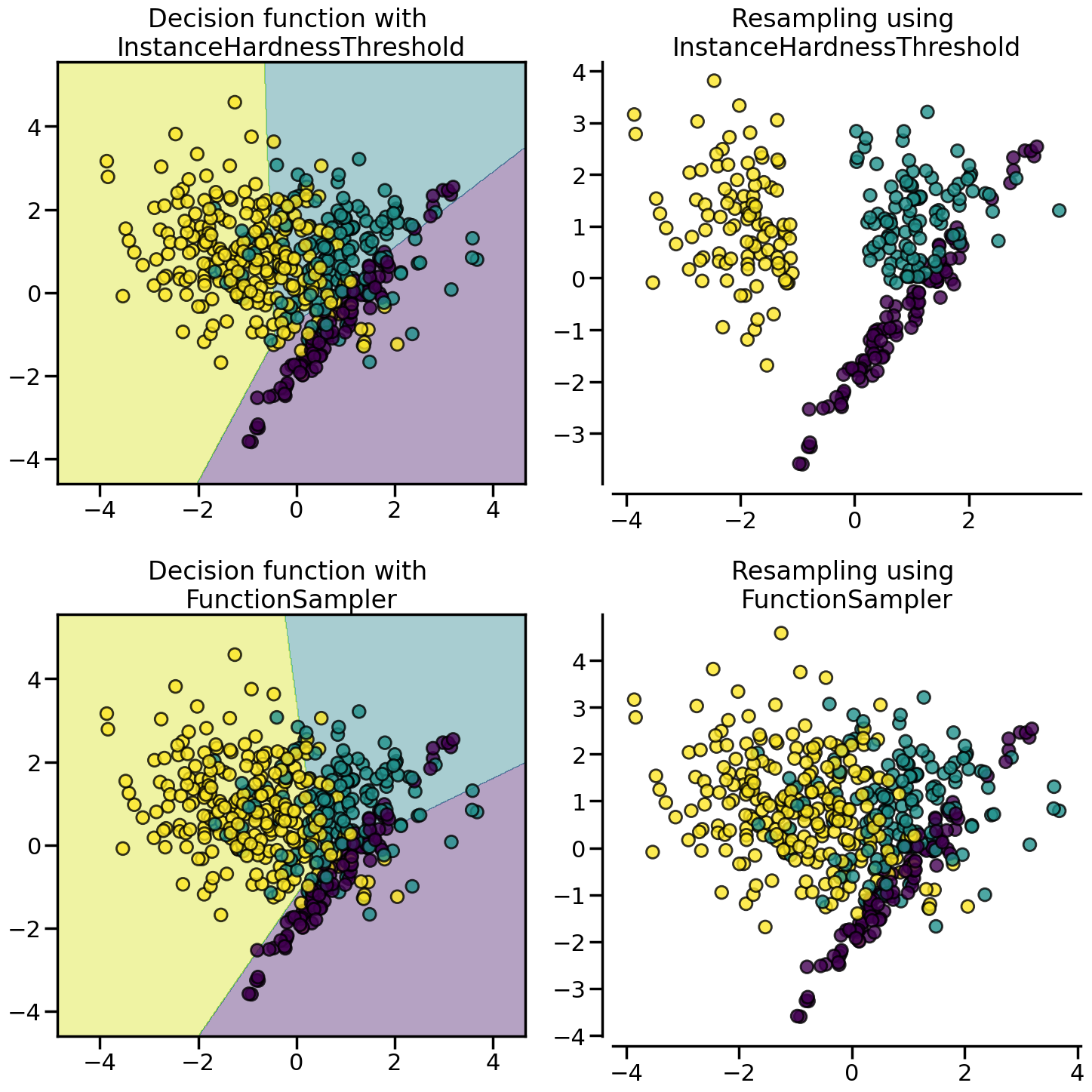
:class:~imblearn.under_sampling.InstanceHardnessThreshold uses the prediction of classifier to exclude samples. All samples which are classified with a low probability will be removed.
from imblearn.under_sampling import InstanceHardnessThreshold
samplers = {
FunctionSampler(), # identity resampler
InstanceHardnessThreshold(
estimator=LogisticRegression(),
random_state=0,
),
}
fig, axs = plt.subplots(nrows=2, ncols=2, figsize=(15, 15))
for ax, sampler in zip(axs, samplers):
model = make_pipeline(sampler, clf).fit(X, y)
plot_decision_function(
X,
y,
model,
ax[0],
title=f"Decision function with \n{sampler.__class__.__name__}",
)
plot_resampling(
X, y, sampler, ax[1], title=f"Resampling using \n{sampler.__class__.__name__}"
)
fig.tight_layout()
plt.show()